Reconsideration Reproducibility of Currently Deep Learning-Based Radiomics: Taking Renal Cell Carcinoma as an Example [Preprint]
Reconsideration Reproducibility of Currently Deep Learning-Based Radiomics: Taking Renal Cell Carcinoma as an Example [Preprint]
Teng Zuo1, Lingfeng He2, Zezheng Lin3, Jianhui Chen4* and Ning Li1*
1 China Medical University
2 Institute for Empirical Social Science Research, Xi'an Jiaotong University
3 United Nations Industrial Development Organization (UNIDO)
4 Fujian Medical University - Fujian Medical University Union Hospital
Computer science and hardware have developed prominently in this decade, advancing Artificial Intelligence and Deep Learning applications in translational medicine. As an icon, DL-radiomics research mushrooms and solves several traditional radiological challenges. Behind the glory of DL-radiomics successful performance, there is limited attention to the neglected reproducibility of existing reports, which runs contrary to radiomics original intention, to realize unexperienced-dependent radiological processing with high robustness and generalization. Besides focusing on objective causes of reproduction barriers, deep-seated factors, between contemporary academic evaluation systems and scientific research, should also be mentioned. There is an urgent need for a targeted inspection to promote this area’s healthy development. We take Renal cell carcinoma as an example, one of the common genitourinary cancers, to glimpse the reproducibility defects in the whole DL-radiomics field. This study then proposes a reproducibility specification checklist with an analysis of the performance of existing DL-radiomics reports in RCC. The results show a trend of increasing reproducibility but still a need to further improve, especially in technological details of pre-processing, training, validation, and testing.
Keywords: RCC, renal cell carcinoma; DL, deep learning; radiomics; reproducibility
Declaration of ethics approval and consent to participate Not applicable.
Declaration of consent for publication Not applicable.
Declaration of competing interests The authors declare that there are no competing interests.
Citation Zuo, T., He, L., Lin, Z., Chen, J., & Li, N. (2023). Reconsideration Reproducibility of Currently Deep Learning-Based Radiomics: Taking Renal Cell Carcinoma as an Example. (SSRN Scholarly Paper No. 4435866). https://doi.org/10.2139/ssrn.4435866
Abbreviations
| DL | Deep Learning |
| RCC | Renal Cell Carcinoma |
| AI | Artificial Intelligent |
| ML | Machine Learning |
| ccRCC | Clear Cell Renal Cell Carcinoma |
| nccRCC | Non-clear Cell Renal Cell Carcinoma |
| pRCC | Papillary Renal Cell Carcinoma |
| chRCC | Chromophobe Renal Cell Carcinoma |
| SRMs | Small Renal Masses |
| RTB | Renal Tumor Biopsy |
| CV | Computer Vision |
| ILSVRC | ImageNet Large Scale Visual Recognition Challenge |
| STEM | Science, Technology, Engineering, and Mathematics |
| CLIAM | Checklist for Artificial Intelligent in Medicine |
| RQS | Radiomics Quality Assessment |
| GAN | Generative Adversarial Network-GAN, |
| DLRRC | Deep Learning Radiomics Reproducibility Checklist |
| MeSH | Medical Subject Headings |
| PRISMA | Preferred Reporting Items for Systematic reviews and Meta-Analyses |
Introduction
Radiomics, aiming to promote understanding of medical imaging by extracting complex features from large datasets (Gillies et al., 2016), is a widely discussed field in translational medicine and digital medicine. Going with the tide of historical development of medical Artificial Intelligent(AI), methods of radiomics have been replicated several times (Hosny et al., 2018). Now, the mainstream of methods involves Machine Learning (ML) and Deep Learning (DL), defined by disparate workflows. Despite extensive research has shown the superiority of DL-radiomics comparing with ML-radiomics while processing large-scale datasets and DL-radiomics widely applied in medical imaging processing (Cruz-Roa et al., 2017; Litjens et al., 2017), DL-related translation into clinical application does not happen yet.
The responsibility of DL-radiomics had captured the concern of academia, especially after the sudden outbreak of COVID-19 and following mushrooming of DL applications in this field (Hryniewska et al., 2021). Nowadays, it is not unusual to witness article retractions in the field of DL-radiomics (Hu et al., 2022; Ma & Jimenez, 2022; Mohammed et al., 2021). In this stage, reproducibility, as the bedrock of authenticity and translation, should be focused on and applied to targeted reviewing.
For a better understanding of this problem, we choose Renal Cell Carcinoma (RCC), one of the main subtypes of genitourinary oncology, as an example to analyse the deeper layers of the reproducibility concept. In this perspective, we took RCC-related research as examples, summarized the issues of research reproducibility, presented factors of reproducibility, analyse the sharp contradiction between the modern evaluation systems and the nature of scientific research as a root cause, provide feasible measures under existing ethical reviewing structure, and evaluate how to solve main challenges in DL-radiomics and move forward.
The Need and Advances of DL-Radiomics in RCC
Renal cell carcinoma (RCC) is one of the main subtypes of genitourinary oncology, with more than 300,000 new cases diagnosed each year (Ferlay et al., 2015). Driven by various mechanisms and genes, RCC includes several subtypes, which were normally divided into two categories based on microscopic features, clear cell RCC (ccRCC) and non-clear cell RCC (nccRCC). After the recent advancement in pathological and genetical knowledge, nccRCC are further subdivided into several classes, including papillary RCC (pRCC), chromophobe RCC (chRCC), and some rare subtypes. Molecular pathology development has propelled the understanding of biological behavior driving mechanisms and given birth to the further molecular classifications, which forebodes the precision diagnosis and treatment age coming.
From a retroperitoneal organ, signs of RCC are mostly asymptomatic or nonspecific, which classic triad of hematuria, pain, and mass occurs 5~10% (Rini et al., 2009). What is worse, due to anatomical structure of kidney and adjacent tissues, it is hard to detect RCC through physical examinations while masses are small. Additionally, some natural history of RCC is variable, even can be asymptomatic. In the clinic, a substantial portion of firstly-diagnosed patients is informed by unintentional radiological examnintaion.
Recently, increasing use of radiology in treatment and diagnosis probably cause incidents to rise in many countries (M. Sun et al., 2011; Y. Yang et al., 2013; Znaor et al., 2015), which finally cause an increasing concern of RCC radiological processing. However, the performance of subjective radiological interpretation is imperfect (Hindman et al., 2012; X.-Y. Sun et al., 2020), especially in differentiation of subtypes (Rossi et al., 2018).
Certain renal tumor subtypes have specific diagnosable characteristics (Diaz de Leon & Pedrosa, 2017), like predominantly cystic mass with irregular and nodular septa of low-grade ccRCC (Pedrosa et al., 2008). In traditional ideas, three main types of RCC have classical diagnostic characteristics, involving hypervascular & hypovascular, vairous peak enhancement during different phases (Lee-Felker et al., 2014; M. R. M. Sun et al., 2009; Young et al., 2013; Zhang et al., 2007). Other studies have also discovered several imaging features correlated with high-grade tumors (Mileto et al., 2014; Pedrosa et al., 2008; Rosenkrantz et al., 2010; Roy et al., 2005). However, the imaging characteristics of RCC are highly variable (Diaz de Leon & Pedrosa, 2017), especially in small renal masses(SRMs). For SRMs, which are smaller than 4cm and usually detected by radiological imaging incidentally (Gill et al., 2010), available radiological technology can’t distinguish, with high confidence, the different subtypes. For example, to differentiate from ccRCC, pRCC can be detected with intralesional hemorrhage (Diaz de Leon & Pedrosa, 2017). However, in SRMs and some undersized nontypical renal masses, the hemorrhage isn’t always observable. If this patient can’t finish renal tumor biopsy(RTB), it will be quite passive for clinicians to proceed to following treatment. In this stage, improvement of radiology would be more suitable for promoting diagnostic performance of RCC. There is an urgent need for a non-empirical quantitative measure.
In the past decade, electronic computer technology rapidly development, like nanomete process changes from 2012 (Nvidia GK104 28nm) to 2022 (Nvidia AD102 5nm) and accompanying calculate performance improvement, provided the base of large-scale calculating, which fits the need of deep neural network, the typical DL algorithm. Since 2012, the evolution of DL in computer vision (CV) has coming because of AlexNet surpassed performance in ImageNet Large Scale Visual Recognition Challenge (ILSVRC) (Krizhevsky et al., 2017). Besides, high profits and growths of DL have attracted hardware and software manufacturers to involve in dependent environment development, prompting lowering accessibility threshold. Now, it isn’t hard to deploy a DL model in a computer with TensorFlow/PyTorch and Nvidia/Intel hardwares for a new-comer in this field. DL-radiomics, with higher performance and lower barriers, has became a heavily discussed topic nowadays. It has shown impressive power in various tasks of RCC, like prognosis via classification (Ning et al., 2020) and treatment selection via detection (Toda et al., 2022), which are believed to solve radiological challenges and promote efficiency.
Limited by incidence rate of RCC and insufficient data size of RCC’s opening imaging source, most reports in RCC DL-Radiomics are stereotypical based on restricted cases. Nonetheless, this field still involves several primary tasks, including classification (Coy et al., 2019; Nikpanah et al., 2021; Oberai et al., 2020), segmentation (Z. Lin et al., 2021) and detection (Islam et al., 2022). Now, several traditional radiological challenges had been resolved, like the differentiation of RCC subtypes and segmentation of blurry boundary. With an admirable performance, RCC DL-Radiomics are believed with high potential to promote radiological diagnosis efficiency.
Reproducibility, the Main Barrier in Translation
As an emerging field in radiology, DL-radiomics now is widely applied in almost every area of medicine research, not only in RCC. The threshold of DL-radiomics technological barrier decreases, contributing to the techno-explosion in medicine. It is easy to forecast the bright future of DL-Radiomics with high accessibility of required softwares and hardwares currently.
DL-Radiomics, as a heavily-discussed part of translational medicine for a decade, which, disappointingly, leaving developing clinical practice applications of biomedical sciences without adequate discussion in clinical practice. Immediate causes involve the accessibility of codes, data and weights, which are undervalued of existing checklists. Considering manual processes existed in most workflows of existing researches, “black box” training and possible random selection of partitions, it is hardly possible to reproduce a similar result without weights and codes, even with a standard protocol and described processing details. Worsening, there are several excuses that can be used to refuse opening access, like intellectual property and ethical protection. In the eyes of people with malice motivations, DL-Radiomics is a buck of Emmental cheese, holey and delicious.
An emerging research field is usually a hardest hit area of academic controversies without effective standards, which is a common occurrence of modern scientific research and have an inkling in DL-Radiomics. The key player of this abnormal phenomenon is intense contradictions between modern scientific evaluation systems and striving in research, which is the predictable outcome of a neoliberalist academia and its system of knowledge production.
For many years, studies and critics had been in the lamentable state in which the notion “publish or perish” had become the law of the land. The modern academic system of knowledge production, especially that of STEM (Science, technology, engineering, and mathematics) research, is, as Max Weber had so elegantly put it, a system of state capitalistic corporation in which the employer suffers from alienation, institutional pressure of publishing, and constant fear of losing their jobs (Zhou & Volkwein, 2004) or missing out in promotion due to mere fate or even luck (Weber, 1946). As the tenure-track system is implemented, as private universities became more ranking-sensitive and donation-sensitive (Boulton, 2011; Cyrenne & Grant, 2009; Linton et al., 2011), competition turns violent. The number of publications, the value of influential factors (Link, 2015) of each publications, and the sector-place of the journal upon which the publication in SSCI or SCI establishment became the center for the careers of today’s “Quantified Scholars” (Pardo-Guerra, 2022) in today’s system of “Digital Taylorism” (Lauder et al., 2008). With more high-quality publications, come higher positions, better professional repute, and higher chance of attaining more funding or grants (Angell, 1986; De Rond & Miller, 2005), which the modern private academic institutes rely more and more desperately. Rising number of Scientists and limited number of funding or publishing vacancy exacerbated the trend (Bornmann & Mutz, 2015).
The unfortunate prevalence of many predatory journals with exploding prices shows the miles that young scholars are willing to go to cope under such extreme stress (Cuschieri & Grech, 2018). Under such pressure, unfortunately, certain scholars adapt disingenuous representations of their works. Embellishing, salami slicing, almost became too common in Medical publishing and news (Chang, 2015; Owen, 2004). Even worse, the occurrence for ever more rampant academic fraudulence became a predictable vice under such system (Altman & Krzywinski, 2017; Chavalarias et al., 2016; Colquhoun, 2014; Fanelli, 2010; Halsey et al., 2015; Krawczyk, 2015; Neill, 2008). Medical science and Biological science in particular, has now a repetition crisis (Baker, 2016; Salman et al., 2014), with certain estimation of malpractice to be “as common as 75%” (Fanelli, 2009). This fierce competition had even pushed Journals, which are also seeking for more exposure and citations, to publish more novel and distinguished results, pushing many scholars to forge their academic findings (Edwards & Roy, 2017). Studies even found that as “fixation in top-tier journals on significant or positive findings tend to drive trustworthiness down, and is more likely to select for false positives and fraudulent results” (Grimes et al., 2018). The fact of Medical science research are usually conducted by groups of many people, with great number of experiment needed, data produced, only makes verification harder and falsification a lot easier, which added to the risk of falsification occurring.
All this, paints a vivid picture of neoliberalist academia and its inevitable result. Neoliberalism, is, at its core, a system of offering and constantly shifting identities (Ong, 2006), which inspires all participants to drown in a vicious cycle of never-ending cut-throat battle. With apparatuses like Journals, and governing technology like Influential Factor calculator, the global assemblage (Ong & Collier, 2005) of academic cohort as a field of social conflict is formed. The neoliberalist calculation machine, rendering evaluation of each and everyone’s “value”, reducing every one as “bare individuals” being disembedded from their social and political relations (Pabst, 2023), constantly offering and shifting identities to scholars, and encouraging insurgence of social and academic status in a Darwinist fashion, without a feasible verification system in place, will lead to the individuals constantly race to the bottom as they race to the top to occupy more publishing space and exposure. As neoliberalism constantly shift individual identity, it also generates new pressure and incentives for the individual to further participate into this vicious game of “publish or perish”. This system, together with willing or unwilling participants, had formed today’s strange landscape of academic fraudulent that many scholars now thrive on. Because in an age where grand academic, Weberian vision had collapsed, a vicious number and power game is all the masses can have to feel meaningful in their lives.
Remolding of academic evaluation systems are not about to happen quickly, but the chaos of academic controversy isn’t tolerated, which requires the improvement of existing checklists. Being similar to the legislation effects to constraint social functioning, checklists are supposed to become a inspectproof net to prevent intentional or unintentional academic controversy. However, the hole of this net is big enough to drill, caused by imperfections of checklists.
Imperfections of Current Reproducibility Reviewing
Certain concerns were voices regarding DL-Radiomics translations (Kelly et al., 2022), the process from codes to clinic. Reviewing existing reports, especially targeting in quality assessments, usually uses two checklists, CLIAM (Checklist for Artificial Intelligent in Medicine) and RQS (Radiomics Quality Assessment). However, several imperfections of these checklists are obvious, escaping academic attention due to the lack of interdisciplinary background. We summarize the reproducibility-relative clauses of them (Table 1).
Table 1 Reproducibility-related clauses in CLIAM.
| No. | Item |
|---|---|
| 1 | Identification as a study of AI methodology, specifying the category of technology used (eg, deep learning) |
| 3 | Scientific and clinical background, including the intended use and clinical role of the AI approach |
| 4 | Study objectives and hypotheses |
| 6 | Study goal, such as model creation, exploratory study, feasibility study, noninferiority trial |
| 7 | Data sources |
| 9 | Data preprocessing steps |
| 10 | Selection of data subsets, if applicable |
| 13 | How missing data were handled |
| 14 | Definition of ground truth reference standard, in sufficient detail to allow replication |
| 15 | Rationale for choosing the reference standard (if alternatives exist) |
| 16 | Source of ground truth annotations; qualifications and preparation of annotators |
| 20 | How data were assigned to partitions; specify proportions |
| 21 | Level at which partitions are disjoint (eg, image, study, patient, institution) |
| 22 | Detailed description of model, including inputs, outputs, all intermediate layers and connections |
| 23 | Software libraries, frameworks, and packages |
| 24 | Initialization of model parameters (eg, randomization, transfer learning) |
| 25 | Details of training approach, including data augmentation, hyperparameters, number of models trained |
| 26 | Method of selecting the final model |
| 27 | Ensembling techniques, if applicable |
| 28 | Metrics of model performance |
| 32 | Validation or testing on external data |
| 33 | Flow of participants or cases, using a diagram to indicate inclusion and exclusion |
| 35 | Performance metrics for optimal model(s) on all data partitions |
| 36 | Estimates of diagnostic accuracy and their precision |
| 37 | Failure analysis of incorrectly classified cases |
| 40 | Registration number and name of registry |
| 41 | Where the full study protocol can be accessed |
CLIAM inspects the whole processes of DL-radiomics, from data collection to testing, However, several issues are existed:
- Lack of quantitative evaluation. It doesn’t grade the work, which means the eligible boundary is unset. It is unable to provide valid assessment a targeted work or massive hunting.
- Lack of accessibility review in weights and datasets. It can be found in No.41 that CLIAM requires a possible accessibility of codes. Reports are hard to reproduce and to assess authenticity only with the accessibility of codes and described details of processes.
- Weighted in protocol normative comparatively, with limited attention to reproducibility-authenticity details. There are contents with great length existing in CLIAM associated with protocol normalization, which is understandable considering the standardizing requirement as a checklist. There should be some rules in the checklist for further promising of reproducibility and authenticity. It isn’t hard to conceal defects intentionally or unwittingly under the supervision of CLIAM.
RQS 2.0 (Lambin et al., 2017), with significant variations of clauses, is accessible through the following webpage (https://www.radiomics.world/rqs2/dl). Although RQS has a wider scope of examination, it also has some issues existed while applying in reproducibility assessment.
- Contains unreasonable weights of options. Comparing with CLIAM, RQS 2.0 can score the quality of reports. However, the score of some options are not quite matching with their tangible impacts. For example, in the clause of “The algorithm, source code, and coefficients are made publicly available. Add a table detailing the different versions of software & packages used.”, the option of Yes only score for 1 point with 1.64%. It is easy to reproduce with codes and coefficients, which is usually called weights in DL. It is not reasonable to score just 1 point.
- Can be considered too broad and shallow. Original intention of RQS probably is forward-looking quality assessment, which cause RQS that has a too broad scope to assess. It also causes the limited depth of detail analysis, which leads to a non-ideal situation of reproducibility assessments.
It is easy to figure that an urgent need of specific reproducibility-based reviewing is existed in DL-Radiomics. As an emerging field in digital medicine, the focus on reproducibility and standardization at an early stage is believed to promote healthy development.
The main hinder of targeted reviewing is actually coming from the academic ethical requirement. This is not a criticism to ethical reviewing, but an approval, on the contrary. Lowering ethical requirements would be an avalanche, probably causing uncoverable tough situation in academia. Considering the constantly technical development in DL, it could be foreseen that possibility of raising ethical requirements would be existed in future, like Generative Adversarial Network (GAN) and its generability-related privacy disclosures. The academic rigor gives people with ulterior motives a leg up on deceiving, which is undesirable. Sheltering by ethics and intellectual property, people can refuse opening access to codes, datasets and weights, which is similar to piping of dams. The most feasible method to plug is deepening non-sensitive detail requirements, which is the motive to design the new checklist.
A New Checklist, Deep Learning Radiomics Reproducibility Checklist (DLRRC)
To fill the gap, we proposed a new checklist Deep Learning Radiomics Reproducibility Checklist (DLRRC) (Table 2). The particulars of DLRRC and reviewing results of RCC DL-Radiomics studies (Supplement 1) are attached.
Table 2 Deep-Learning Radiomics Research Checklist (DLRRC). This checklist scores 100 points, regarding 50 points as a baseline of “acceptable reproducibility”. The website of DLRRC is https://apps.gmade-studio.com/dlrrc.
| No. | Item with Answers & Scores | ||
|---|---|---|---|
| Part I: Basic information and Data Acquisition (10 points) | |||
| 1 | Labels are meaningful and biological discrepancy, mentioning potential topological differences existing. Answers and scores: Yes (2.5); No (0). | ||
| 2 | Filtration of radiological data are applied and described. Answers and scores: Yes (2.5); No (0). | ||
| 3 | Radiological data types: - If several modalities are involved, each type of modality should have an acceptable ratio with detailed descriptions. - If only one modality is involved, declaration is required. Answers and scores: Yes (2.5); No (0). | ||
| 4 | For data sources: - If it is all originated from open sources, situation of application and filters should be declared. - If it involves closed-source data, institutional ethical reviewing approvement and serial numbers are required. Answers and scores: Yes (2.5); No (0). | ||
| Part II: Pre-processing (27.5 points) | |||
| 5 | The ratio of each label should be unextreme and acceptable. Answers and scores: Yes (2.5); No (0). | ||
| 6 | The ratio of images and cases should be closed, otherwise reasonable explanation is required. Answers and scores: Yes (2.5); No (0). | ||
| 7 | Processing staffs are radiological professionals. Answers and scores: Yes (2.5); No (0). | ||
| 8 | Pre-processing is processed by the gold standard guidance. Answers and scores: Yes (2.5); No (0). | ||
| 9 | Effective methods exist to promise accuracy of pre-processing. Answers and scores: Yes (2.5); No (0). | ||
| 10 | The ratio of each dataset is reasonable. Answers and scores: Yes (2.5); No (0). | ||
| 11 | Cases are independent, which aren’t involved in different datasets. Answers and scores: Yes (2.5); No (0). | ||
| 12 | Datasets are established by random selection, without manual manipulating. Answers and scores: Yes (2.5); No (0). | ||
| 13 | Examples of pre-processing are listed. Answers and scores: Yes (2.5); No (0). | ||
| 14 | Methods of data augmentation are suited and correctly applied. Answers and scores: And (5); Or (2.5); Nor (0). | ||
| Part III: Model and Dependence (10 points) | |||
| 15 | Methods to avoid overfitting are applied and described. Answers and scores: Yes (2.5); No (0). | ||
| 16 | Software environment should be listed, like serial numbers of version. Answers and scores: Yes (2.5); No (0). | ||
| 17 | Hardware details should be listed. Answers and scores: Yes (2.5); No (0). | ||
| 18 | Applied models are suited for tasks. Answers and scores: Yes (2.5); No (0). | ||
| Part IV: Training, Validation and Testing (27.5 points) | |||
| 19 | Training details are listed, like epoch and time spent. Answers and scores: Yes (2.5); No (0). | ||
| 20 | Hyperparameters are listed (at least including batch size and learning rate). Answers and scores: Yes (2.5); No (0). | ||
| 21 | The curves of accuracy-epoch and loss-epoch are provided. Answers and scores: Yes (2.5); No (0). | ||
| 22 | The end of training is decided by the performance trends in validation datasets and is described. Answers and scores: Yes (2.5); No (0). | ||
| 23 | Methods to promote robustness are applied. Answers and scores: Yes (2.5); No (0). | ||
| 24 | The details of initial weights are described. Answers and scores: Yes (2.5); No (0). | ||
| 25 | Training workloads are listed. Answers and scores: Yes (2.5); No (0). | ||
| 26 | Objective indexes are reasonable and complete. Answers and scores: And (5); Or (2.5); Nor (0). | ||
| 27 | Comparison between testing performance and manual processing with the same test dataset is provided. Answers and scores: Yes (2.5); No (0). | ||
| 28 | Details of test datasets and speculation results are provided. Answers and scores: Yes (2.5); No (0). | ||
| Part V: Accessibility (25 points) | |||
| 29 | Codes. Answers and scores: Fully accessible (10); Partly accessible (7.5); Possible accessible (5); Not (0). | ||
| 30 | Datasets. Answers and scores: Fully accessible (10); Partly accessible (7.5); Possible accessible (5); Not (0). | ||
| 31 | Weights. Answers and scores: Accessible (5); Inaccessible (0). | ||
To be noticed, DLRRC is designed for targeted reproducibility assessment of DL-Radiomics, which causes different focuses comparing with CLIAM/RQS. The principal index is reproducibility of reports, which manifest different items. It could be found that our checklist involves clauses mentioned by RQS/CLIAM, and some new requirements. These new requirements, like spent time of each epoch and matched-degree of models & tasks, are applied to profile the authenticity and reproducibility logically. For example:
- We require authors to provide hardware details, parameters of models, datasets size and spent time in each epoch, which can be used to deduce bidirectionally. The expected computing scale and hardware performance will have to spend more time. Also, the computing scale, formed by parameters and datasets size, can be speculated by spent time and hardware performance. This logic is used to assess the authenticity and provide suggestions in reproduction.
- We require authors to provide hardware and software details, involving dependent software version and hardware version, like the versions of TensorFlow/ PyTorch/ CUDA/ CUDNN and graphic card types. There are relevance existed between TensorFlow/PyTorch and CUDA/CUDNN, which means the models can’t perform with inconsistent versions of these dependencies. In some retracted articles, the authenticity can be argued quickly by checking these details. Also, in reproduction, these details are important to deploy models.
There are several veins hiding in this checklist, weaving a blanket over researches in this field. We don’t explain reasons of every clause, but each one equally have an important role in reproducibility assessments. This checklist can be applied in other DL-Radiomics fields as the generalization of DL methods, which can be proved by the scores of retracted articles from different fields of DL-radiomics.
DLRRC Practice and Discovery in RCC
To obtain a glimpse of the current situation about reproducibility of DL-Radiomics, we collect documents of RCC DL-Radiomics from PubMed and Web of Science with certain Medical Subject Headings (MeSH), involving “Neural Networks, Computer”[Mesh], “Deep Learning”[Mesh] and “Carcinoma, Renal Cell”[Mesh]. Finally, 22 peer-reviewed journal articles in RCC field fall in the scope (Baghdadi et al., 2020; Coy et al., 2019; Guo et al., 2022; Han et al., 2019; Hussain et al., 2021; Islam et al., 2022; F. Lin et al., 2020; Z. Lin et al., 2021; Nikpanah et al., 2021; Oberai et al., 2020; Pedersen et al., 2020; Sudharson & Kokil, 2020; Tanaka et al., 2020; Toda et al., 2022; Xi et al., 2020; Xia et al., 2019; Xu et al., 2022; G. Yang et al., 2020; Zabihollahy et al., 2020; Zhao et al., 2020; Zheng et al., 2021; Zhu et al., 2022) and are examined by DLRRC (more details are attached in Supplementary Information).
Figure 1 presents a scatter of JIF percentile via DLRRC scores of the reports. Setting 50 points as the threshold of “acceptable reproducibility”, this study finds that overall quality of RCC DL-Radiomics is relatively acceptable, according to Figure 1. Most reports (i.e., 16 of 22) are partly reproducible, which means similar results can be performed with semblable protocols. Given further analysis, we find that reports with disquieting scores are published earlier than 2021 in some technology-focused journals (c.f., Supplementary Information), which pay more attention to innovations of DL methodology and less attention to overall normalization. Hence, it is encouraged to keep an attention balance between innovations and protocol normalization, which is more reasonable.
Grounded on the above findings, we further analyse the correlation between journal levels and DLRRC scores (Figure 1) and the trend over time (Figure 2). Figure 1 reveals that there is no linear correlation between the journal levels and DLRRC scores (corr = -0.085, p = 0.707; partial corr = -0.125, p = 0.590), indicating that the need of advancing reproducibility is a general issue across all levels of journals. Fortunately, Figure 2 reveals a moderately positively linear correlation between years of publications and DLRRC scores (corr = 0.402, p = 0.064; partial corr = 0.411, p = 0.064; the p value is reasonably accepted due to the limited sample size), hinting a good omen achieved by the academia and explicable due to time-varying accessibility of DL methods.
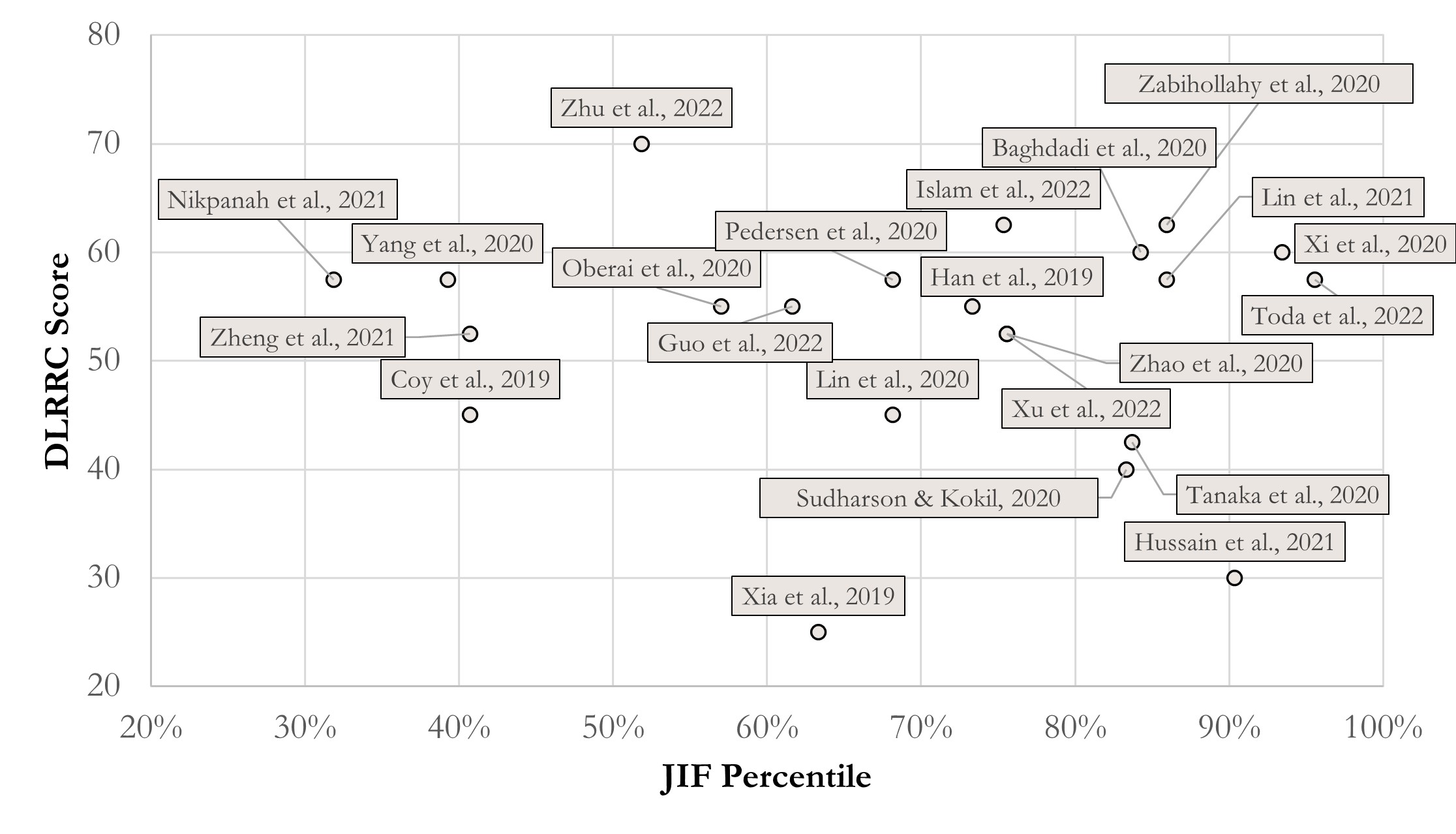
Figure 1 The score scatter of JIF percentile and DLLRC Score in RCC. Correlation coefficients = -0.085 (p = 0.707); partial correlation coefficients = -0.125 (p = 0.590), controlling year effect.
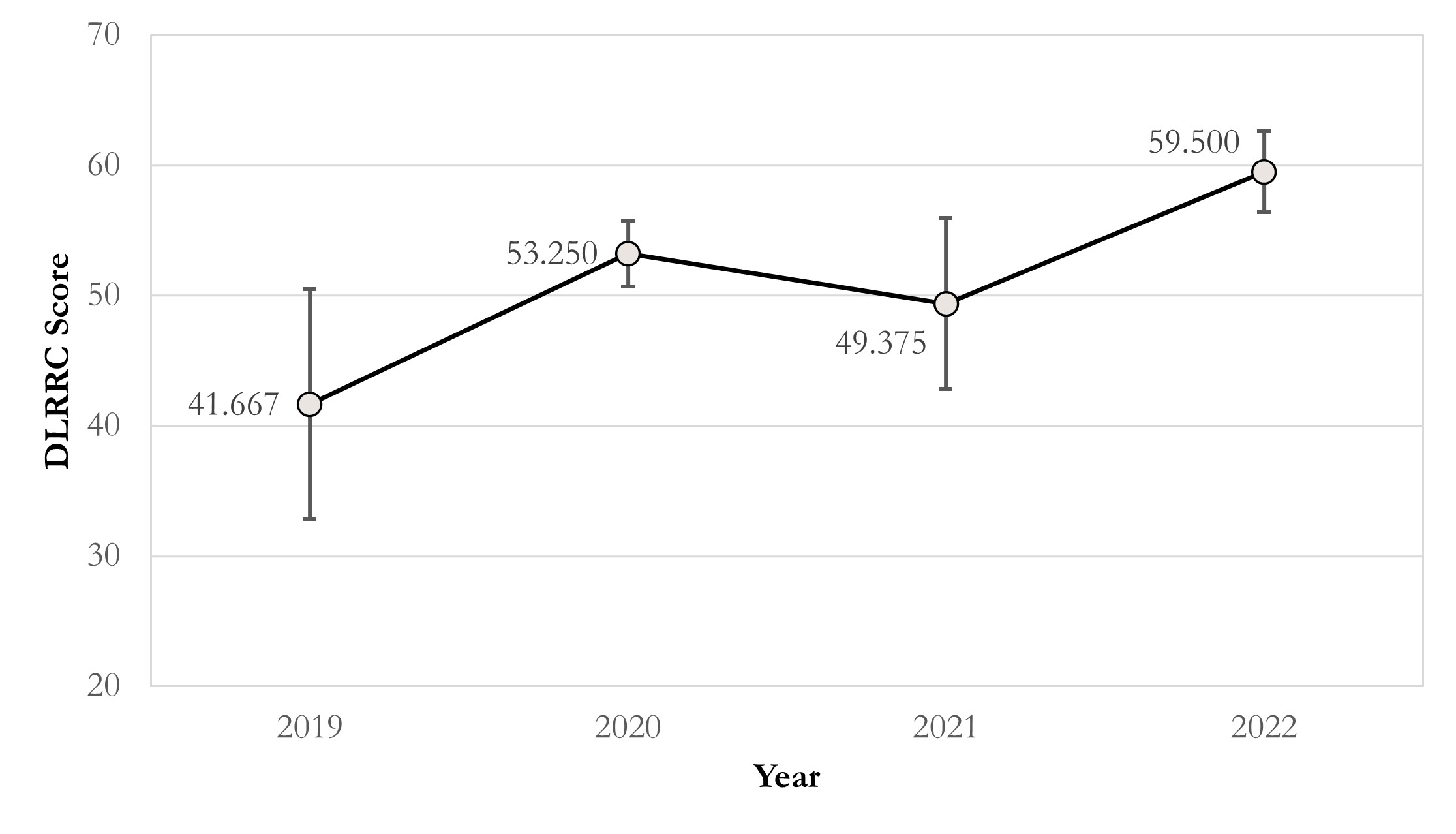
Figure 2 The line chart of DLRRC score in RCC and published year. Correlation coefficients = 0.402 (p = 0.064); partial correlation coefficients = 0.411 (p = 0.064), controlling JIF effect.
Figure 3 presents the average point percentages of each part. The average point percentage of the whole report is 52.4%, equal to the threshold of “acceptable reproducibility”. Part I “Basic information and Data Acquisition” and Part III “Model and Dependence” are relatively well done, with average point percentages of 64.8% and 60.2%, respectively. Part II “Pre-processing” and Part V “Accessibility” are with moderate average point percentages around the threshold of “acceptable reproducibility”. Part IV “Training, Validation and Testing” should be noticed, due to its average point percentage as 42.6%.
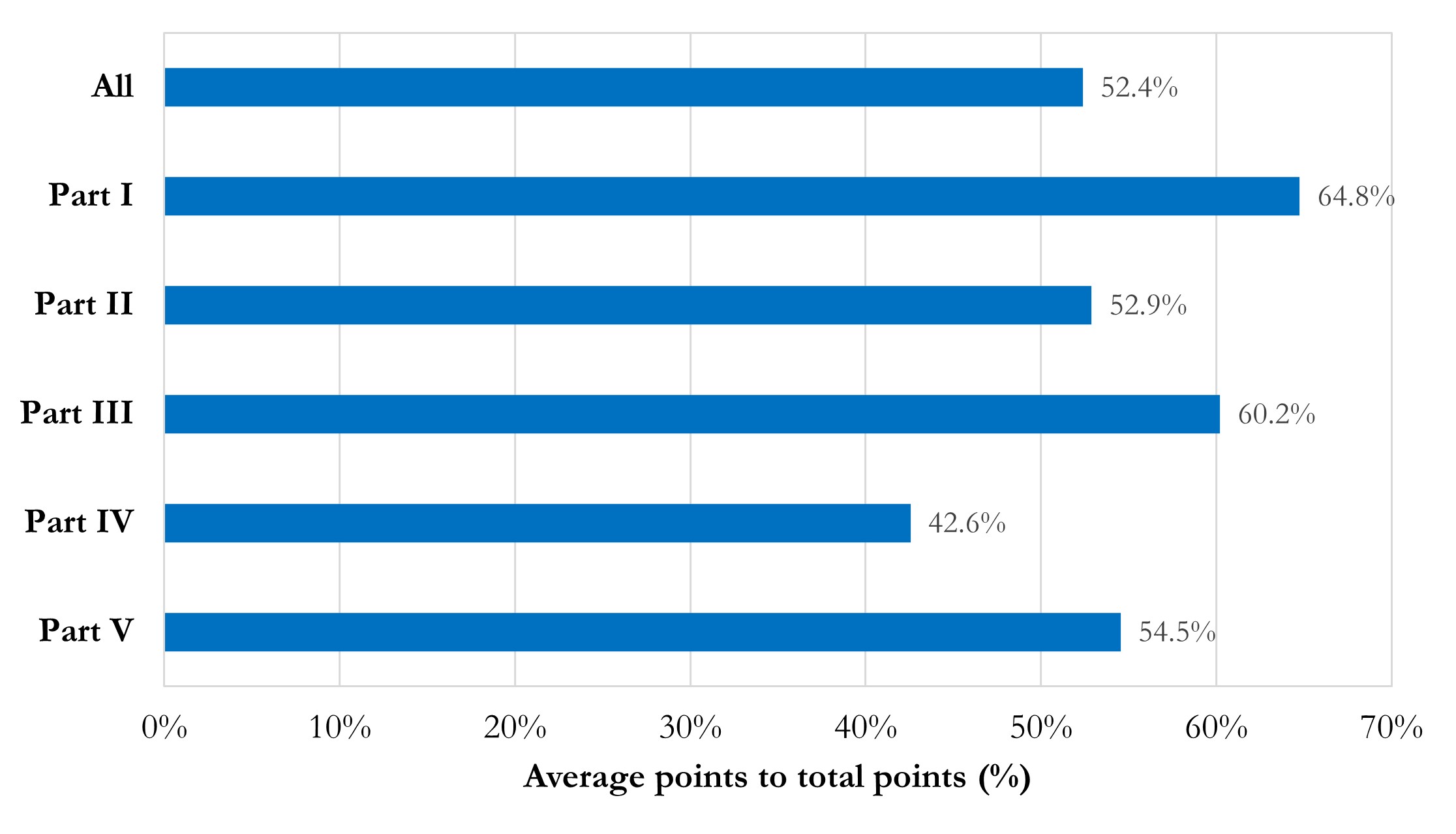
Figure 3 Bar chart of average point percentages in total and each part.
To explore current vulnerability in terms of reproducibility, we summarize the average point percentages of each item in Figure 4. Several discoveries with recommendations are highlighted:
- Accessibility needs improvement. The main discovery is that the impressive weights (item 31 rated 4.5%) and overall accessibility is quite low. Considering possible factors of closed sources existed, we set a baseline of accessibility, which should have 50% points or higher points of Part V scores. It is tolerable that details of models and datasets are described while codes and datasets are non-open and belong to some ongoing projects. Even so, the pass rate of accessibility is still fallacious in general.
- Institutional reviewing number of closed source data should be offered (item 4 rated 18.2%). Some authors declared that the written informed consent was waived and didn’t provide institutional reviewing number, which is not encouraged in our perspective. Usually, as a retrospective study, it truly can be waived of written informed consent. We encourage authors to provide reviewing approval details, as an authenticity evidence of research and inevitable information if authors really register for a retrospective study in institutions.
- Data pre-processing needs standardization to prove robustness. The robustness of results is the cornerstone of reproducibility and comes from standard and well-designed pre-processing and methodology. Unfortunately, issues of data pre-processing are common in established reports. Unbalanced labels’ ratios without suited models (item 5 rated 31.8%), inconsistent ratios of images generation (item 6 rated 27.3%), the lack of referring golden standard guideline during pre-processing (item 8 rated 18.2%), the lack of cross-validation (item 9 rated 31.8%), and manual manipulating during datasets division (item 12 rated 45.5%) may weaken the robustness of results. In addition, it should be noted that random division of training, validation and testing datasets need to be in the patients scale instead of images scale. Otherwise, it may lead to a situation where images of one patient are involved in different datasets, and thus obviously mistaking.
- Technological information on model and dependence, especially software dependencies, should be elaborately attached. Software environment descriptions are often incomplete in existing reports (item 16 rated 4.5%, i.e., only 1 report passing). Versions of CUDA, cuDNN, as well as the deep-learning framework such as TensorFlow and PyTorch are supposed to be listed, because it’s believed that the dependency across those versions can improve the level of confidence about the authenticity and reproducibility of a certain report. However, most authors don’t describe clearly about dependent software environment. Instead, they usually tend to describe ambiguous information, like “using PyTorch” or “with TensorFlow”, much less to more detailed information like applied package version. For example, cuDNN version is declared while CUDA version and TensorFlow/Python version are not declared, which is quirky. It is beyond understanding, considering that no barrier exists in acquisition of these information in a real study.
- Details of training, validation, and testing need to supplement. Except item 26, items of Part IV are rated low.
- Training details about epoch and time spent are hardly listed in the reports (item 19 rated 31.8%); accuracy / loss – epoch curves are rarely presented (item 21 rated 18.2%); and none of studies report the workloads during training (item 25 rated 0%). These details are important, given that they have a potential combination with hardware information. This is understandable as it is not a common index in existing research, especially in non-special application study that rely on edge calculation or low-performance hardware. We encourage authors to provide these details for a better value assessment.
- No declaration of hyper-parameters (item 20 rated 59.1%) and initial weights (item 24 rated 54.5%) is also common in these reports. Again, these details are not technical sensitive, which should have no barrier to acquire in research and declared in articles. We suggest the authors provide non-technical sensitive details as much as possible, for better reproducibility and authenticity.
- Only a few of studies have employed methods for more convincing results (item 22 rated 18.2%, item 23 rated 50.0%). We encourage authors to apply robustness promotion methods, like x-fold cross-validation in model training without a specific validation dataset and determinate training end by the performance trends in validation datasets.
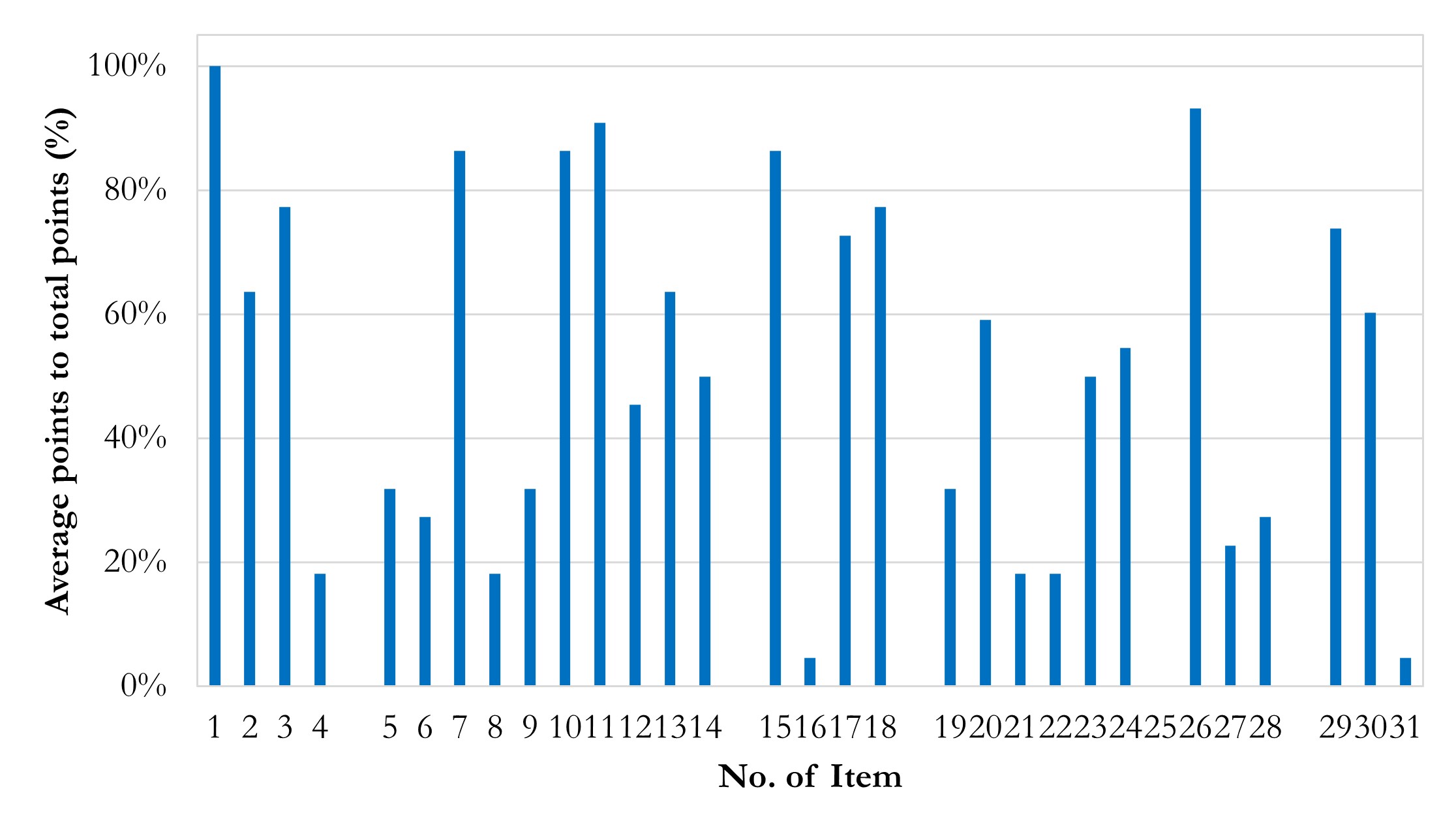
Figure 4 Column chart of average points in items. Average points in Item 16 and 31 are impressively low. Average points in Item 1 are quite high.
Based on the above findings, several implements can be proposed. As the deficiency of DL-Radiomics reports in RCC exists, we call for a widely recognized DL protocol or a detailed guideline to direct following efforts. Furthermore, and significantly, given that article structures are various, influencing readability and detail extraction, we call for an appropriate standard to promote readability by guiding structures of articles, like PRISMA of Meta-analysis. Finally, for a more comprehensive analysis of reproducibility, a wider reviewing of DL-Radiomics reports is required.
DLRRC, as a new generalized checklist, needs more extensive testing and evaluation for proving efficiency. We provide a web application for online assessment with DLRRC (https://apps.gmade-studio.com/dlrrc). Everyone is welcome to use this checklist to assess the reproducibility of DL-Radiomics reports and send their feedback or suggestions. It is too far to say that we hope this perspective and DLRRC can fix these reproducibility defects in DL-Radiomics. Also, it is not a criticism to a certain report. Our original intention is to call for attention from academia to focus on the current situation. We truly hope that the future of DL-Radiomics can be brighter with industry-wide attempts.
Conclusions
We take DL-Radiomics applications in RCC as an example to analyze reproducibility, glimpsing the reproducibility of the whole DL-Radiomics. It is not surprised that mostly reports can’t reproduce completely, as the reproducibility deficiency has been notorious for decades in translational medicine. The current situation is still frustrating. However, scant attention from academia is devoted to this, which is the main motive of this perspective. We truly hope that more practitioners will devote into the healthy development of DL-Radiomics in the future, for a greater tomorrow of translational medicine.
Acknowledgements
We thank all colleagues at Gmade Studio for related discussions.
Availability of Data and Materials
All data can be availed by contacting Gmade Studio(gmadestudio@163.com) and Mr. Teng Zuo.
Author Contributions
Teng Zuo: Conceptualization, Investigation, Writing - Original Draft, Writing - Review & Editing; Lingfeng He: Investigation, Visualization, Writing - Original Draft, Writing - Review & Editing; Zezheng Lin: Writing - Original Draft, Writing - Review & Editing; Jianhui Chen: Writing - Review & Editing, Supervision, Funding acquisition; Ning Li: Writing - Review & Editing, Supervision. All authors read and approved the final manuscript.
Funding information
Teng Zuo and Jianhui Chen were supported by the Training Program for Young and Middle-aged elite Talents of Fujian Provincial Health Commission (2021GGA014).
Role of the funding source The funding source(s) had no involved in the research design.
References
Altman, N., & Krzywinski, M. (2017). P values and the search for significance. Nature Methods, 14(1), 3–4. https://doi.org/10.1038/nmeth.4120
Angell, M. (1986). Publish or Perish: A Proposal. Annals of Internal Medicine, 104(2), 261. https://doi.org/10.7326/0003-4819-104-2-261
Baghdadi, A., Aldhaam, N. A., Elsayed, A. S., Hussein, A. A., Cavuoto, L. A., Kauffman, E., & Guru, K. A. (2020). Automated differentiation of benign renal oncocytoma and chromophobe renal cell carcinoma on computed tomography using deep learning. BJU Int, 125(4), 553–560. https://doi.org/10.1111/bju.14985
Baker, M. (2016). Is there a reproducibility crisis? Nature, 533(7604), 452–454. https://doi.org/10.1038/533452a
Bornmann, L., & Mutz, R. (2015). Growth rates of modern science: A bibliometric analysis based on the number of publications and cited references: Growth Rates of Modern Science: A Bibliometric Analysis Based on the Number of Publications and Cited References. Journal of the Association for Information Science and Technology, 66(11), 2215–2222. https://doi.org/10.1002/asi.23329
Boulton, G. (2011). University Rankings: Diversity, Excellence and the European Initiative. Procedia - Social and Behavioral Sciences, 13, 74–82. https://doi.org/10.1016/j.sbspro.2011.03.006
Chang, C. (2015). Motivated Processing: How People Perceive News Covering Novel or Contradictory Health Research Findings. Science Communication, 37(5), 602–634. https://doi.org/10.1177/1075547015597914
Chavalarias, D., Wallach, J. D., Li, A. H. T., & Ioannidis, J. P. A. (2016). Evolution of Reporting P Values in the Biomedical Literature, 1990-2015. JAMA, 315(11), 1141. https://doi.org/10.1001/jama.2016.1952
Colquhoun, D. (2014). An investigation of the false discovery rate and the misinterpretation of p -values. Royal Society Open Science, 1(3), 140216. https://doi.org/10.1098/rsos.140216
Coy, H., Hsieh, K., Wu, W., Nagarajan, M. B., Young, J. R., Douek, M. L., Brown, M. S., Scalzo, F., & Raman, S. S. (2019). Deep learning and radiomics: The utility of Google TensorFlowTM Inception in classifying clear cell renal cell carcinoma and oncocytoma on multiphasic CT. Abdominal Radiology (New York), 44(6), 2009–2020. https://doi.org/10.1007/s00261-019-01929-0
Cruz-Roa, A., Gilmore, H., Basavanhally, A., Feldman, M., Ganesan, S., Shih, N. N. C., Tomaszewski, J., González, F. A., & Madabhushi, A. (2017). Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent. Scientific Reports, 7(1), 46450. https://doi.org/10.1038/srep46450
Cuschieri, S., & Grech, V. (2018). WASP (Write a Scientific Paper): Open access unsolicited emails for scholarly work – Young and senior researchers perspectives. Early Human Development, 122, 64–66. https://doi.org/10.1016/j.earlhumdev.2018.04.006
Cyrenne, P., & Grant, H. (2009). University decision making and prestige: An empirical study. Economics of Education Review, 28(2), 237–248. https://doi.org/10.1016/j.econedurev.2008.06.001
De Rond, M., & Miller, A. N. (2005). Publish or Perish: Bane or Boon of Academic Life? Journal of Management Inquiry, 14(4), 321–329. https://doi.org/10.1177/1056492605276850
Diaz de Leon, A., & Pedrosa, I. (2017). Imaging and Screening of Kidney Cancer. Radiologic Clinics of North America, 55(6), 1235–1250. https://doi.org/10.1016/j.rcl.2017.06.007
Edwards, M. A., & Roy, S. (2017). Academic Research in the 21st Century: Maintaining Scientific Integrity in a Climate of Perverse Incentives and Hypercompetition. Environmental Engineering Science, 34(1), 51–61. https://doi.org/10.1089/ees.2016.0223
Fanelli, D. (2009). How Many Scientists Fabricate and Falsify Research? A Systematic Review and Meta-Analysis of Survey Data. PLoS ONE, 4(5), e5738. https://doi.org/10.1371/journal.pone.0005738
Fanelli, D. (2010). Do Pressures to Publish Increase Scientists’ Bias? An Empirical Support from US States Data. PLoS ONE, 5(4), e10271. https://doi.org/10.1371/journal.pone.0010271
Ferlay, J., Soerjomataram, I., Dikshit, R., Eser, S., Mathers, C., Rebelo, M., Parkin, D. M., Forman, D., & Bray, F. (2015). Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012: Globocan 2012. International Journal of Cancer, 136(5), E359–E386. https://doi.org/10.1002/ijc.29210
Gill, I. S., Aron, M., Gervais, D. A., & Jewett, M. A. S. (2010). Small Renal Mass. New England Journal of Medicine, 362(7), 624–634. https://doi.org/10.1056/NEJMcp0910041
Gillies, R. J., Kinahan, P. E., & Hricak, H. (2016). Radiomics: Images Are More than Pictures, They Are Data. Radiology, 278(2), 563–577. https://doi.org/10.1148/radiol.2015151169
Grimes, D. R., Bauch, C. T., & Ioannidis, J. P. A. (2018). Modelling science trustworthiness under publish or perish pressure. Royal Society Open Science, 5(1), 171511. https://doi.org/10.1098/rsos.171511
Guo, J., Odu, A., & Pedrosa, I. (2022). Deep learning kidney segmentation with very limited training data using a cascaded convolution neural network. Plos One, 17(5), e0267753. https://doi.org/10.1371/journal.pone.0267753
Halsey, L. G., Curran-Everett, D., Vowler, S. L., & Drummond, G. B. (2015). The fickle P value generates irreproducible results. Nature Methods, 12(3), 179–185. https://doi.org/10.1038/nmeth.3288
Han, S., Hwang, S. I., & Lee, H. J. (2019). The Classification of Renal Cancer in 3-Phase CT Images Using a Deep Learning Method. J Digit Imaging, 32(4), 638–643. https://doi.org/10.1007/s10278-019-00230-2
Hindman, N., Ngo, L., Genega, E. M., Melamed, J., Wei, J., Braza, J. M., Rofsky, N. M., & Pedrosa, I. (2012). Angiomyolipoma with Minimal Fat: Can It Be Differentiated from Clear Cell Renal Cell Carcinoma by Using Standard MR Techniques? Radiology, 265(2), 468–477. https://doi.org/10.1148/radiol.12112087
Hosny, A., Parmar, C., Quackenbush, J., Schwartz, L. H., & Aerts, H. J. W. L. (2018). Artificial intelligence in radiology. Nature Reviews Cancer, 18(8), 500–510. https://doi.org/10.1038/s41568-018-0016-5
Hryniewska, W., Bombiński, P., Szatkowski, P., Tomaszewska, P., Przelaskowski, A., & Biecek, P. (2021). Checklist for responsible deep learning modeling of medical images based on COVID-19 detection studies. Pattern Recognition, 118, 108035. https://doi.org/10.1016/j.patcog.2021.108035
Hu, G., Qian, F., Sha, L., & Wei, Z. (2022). Application of Deep Learning Technology in Glioma. Journal of Healthcare Engineering, 2022, 1–9. https://doi.org/10.1155/2022/8507773
Hussain, M. A., Hamarneh, G., & Garbi, R. (2021). Learnable image histograms-based deep radiomics for renal cell carcinoma grading and staging. Computerized Medical Imaging and Graphics, 90, 101924. https://doi.org/10.1016/j.compmedimag.2021.101924
Islam, M. N., Hasan, M., Hossain, M. K., Alam, M. G. R., Uddin, M. Z., & Soylu, A. (2022). Vision transformer and explainable transfer learning models for auto detection of kidney cyst, stone and tumor from CT-radiography. Scientific Reports, 12(1), 11440. https://doi.org/10.1038/s41598-022-15634-4
Kelly, B. S., Judge, C., Bollard, S. M., Clifford, S. M., Healy, G. M., Aziz, A., Mathur, P., Islam, S., Yeom, K. W., Lawlor, A., & Killeen, R. P. (2022). Radiology artificial intelligence: A systematic review and evaluation of methods (RAISE). European Radiology, 32(11), 7998–8007. https://doi.org/10.1007/s00330-022-08784-6
Krawczyk, M. (2015). The Search for Significance: A Few Peculiarities in the Distribution of P Values in Experimental Psychology Literature. PLOS ONE, 10(6), e0127872. https://doi.org/10.1371/journal.pone.0127872
Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2017). ImageNet classification with deep convolutional neural networks. Communications of the ACM, 60(6), 84–90. https://doi.org/10.1145/3065386
Lambin, P., Leijenaar, R. T. H., Deist, T. M., Peerlings, J., de Jong, E. E. C., van Timmeren, J., Sanduleanu, S., Larue, R. T. H. M., Even, A. J. G., Jochems, A., van Wijk, Y., Woodruff, H., van Soest, J., Lustberg, T., Roelofs, E., van Elmpt, W., Dekker, A., Mottaghy, F. M., Wildberger, J. E., & Walsh, S. (2017). Radiomics: The bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol, 14(12), 749–762. https://doi.org/10.1038/nrclinonc.2017.141
Lauder, H., Brown, P., & Brown, C. (2008). The consequences of global expansion for knowledge, creativity and communication: An analysis and scenario. ResearchGate. https://www.researchgate.net/publication/253764919_The_consequences_of_global_expansion_for_knowledge_creativity_and_communication_an_analysis_and_scenario
Lee-Felker, S. A., Felker, E. R., Tan, N., Margolis, D. J. A., Young, J. R., Sayre, J., & Raman, S. S. (2014). Qualitative and Quantitative MDCT Features for Differentiating Clear Cell Renal Cell Carcinoma From Other Solid Renal Cortical Masses. American Journal of Roentgenology, 203(5), W516–W524. https://doi.org/10.2214/AJR.14.12460
Lin, F., Ma, C., Xu, J., Lei, Y., Li, Q., Lan, Y., Sun, M., Long, W., & Cui, E. (2020). A CT-based deep learning model for predicting the nuclear grade of clear cell renal cell carcinoma. European Journal of Radiology, 129, 109079. https://doi.org/10.1016/j.ejrad.2020.109079
Lin, Z., Cui, Y., Liu, J., Sun, Z., Ma, S., Zhang, X., & Wang, X. (2021). Automated segmentation of kidney and renal mass and automated detection of renal mass in CT urography using 3D U-Net-based deep convolutional neural network. European Radiology, 31(7), 5021–5031. https://doi.org/10.1007/s00330-020-07608-9
Link, J. M. (2015). Publish or perish…but where? What is the value of impact factors? Nuclear Medicine and Biology, 42(5), 426–427. https://doi.org/10.1016/j.nucmedbio.2015.01.004
Linton, J. D., Tierney, R., & Walsh, S. T. (2011). Publish or Perish: How Are Research and Reputation Related? Serials Review, 37(4), 244–257. https://doi.org/10.1080/00987913.2011.10765398
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M., van der Laak, J. A. W. M., van Ginneken, B., & Sánchez, C. I. (2017). A survey on deep learning in medical image analysis. Medical Image Analysis, 42, 60–88. https://doi.org/10.1016/j.media.2017.07.005
Ma, Q., & Jimenez, G. (2022). RETRACTED: Lung cancer diagnosis of CT images using metaheuristics and deep learning. Proceedings of the Institution of Mechanical Engineers, Part H: Journal of Engineering in Medicine, 095441192210907. https://doi.org/10.1177/09544119221090725
Mileto, A., Marin, D., Alfaro-Cordoba, M., Ramirez-Giraldo, J. C., Eusemann, C. D., Scribano, E., Blandino, A., Mazziotti, S., & Ascenti, G. (2014). Iodine Quantification to Distinguish Clear Cell from Papillary Renal Cell Carcinoma at Dual-Energy Multidetector CT: A Multireader Diagnostic Performance Study. Radiology, 273(3), 813–820. https://doi.org/10.1148/radiol.14140171
Mohammed, F., He, X., & Lin, Y. (2021). Retracted: An easy-to-use deep-learning model for highly accurate diagnosis of Parkinson’s disease using SPECT images. Computerized Medical Imaging and Graphics, 87, 101810. https://doi.org/10.1016/j.compmedimag.2020.101810
Neill, U. S. (2008). Publish or perish, but at what cost? Journal of Clinical Investigation, 118(7), 2368–2368. https://doi.org/10.1172/JCI36371
Nikpanah, M., Xu, Z., Jin, D., Farhadi, F., Saboury, B., Ball, M. W., Gautam, R., Merino, M. J., Wood, B. J., Turkbey, B., Jones, E. C., Linehan, W. M., & Malayeri, A. A. (2021). A deep-learning based artificial intelligence (AI) approach for differentiation of clear cell renal cell carcinoma from oncocytoma on multi-phasic MRI. Clinical Imaging, 77, 291–298. https://doi.org/10.1016/j.clinimag.2021.06.016
Ning, Z., Pan, W., Chen, Y., Xiao, Q., Zhang, X., Luo, J., Wang, J., & Zhang, Y. (2020). Integrative analysis of cross-modal features for the prognosis prediction of clear cell renal cell carcinoma. Bioinformatics (Oxford, England), 36(9), 2888–2895. https://doi.org/10.1093/bioinformatics/btaa056
Oberai, A., Varghese, B., Cen, S., Angelini, T., Hwang, D., Gill, I., Aron, M., Lau, C., & Duddalwar, V. (2020). Deep learning based classification of solid lipid-poor contrast enhancing renal masses using contrast enhanced CT. The British Journal of Radiology, 93(1111), 20200002. https://doi.org/10.1259/bjr.20200002
Ong, A. (2006). Neoliberalism as exception: Mutations in citizenship and sovereignty. Duke University Press.
Ong, A., & Collier, S. J. (Eds.). (2005). Global assemblages: Technology, politics, and ethics as anthropological problems. Blackwell Publishing.
Owen, W. J. (2004, February 9). In Defense of the Least Publishable Unit. The Chronicle of Higher Education. https://www.chronicle.com/article/in-defense-of-the-least-publishable-unit/
Pabst, A. (2023, March 8). Why universities are making us stupid. New Statesman. https://www.newstatesman.com/long-reads/2023/03/universities-making-us-stupid
Pardo-Guerra, J. P. (2022). The quantified scholar: How research evaluations transformed the British social sciences. Columbia University Press.
Pedersen, M., Andersen, M. B., Christiansen, H., & Azawi, N. H. (2020). Classification of renal tumour using convolutional neural networks to detect oncocytoma. European Journal of Radiology, 133, 109343. https://doi.org/10.1016/j.ejrad.2020.109343
Pedrosa, I., Chou, M. T., Ngo, L., H. Baroni, R., Genega, E. M., Galaburda, L., DeWolf, W. C., & Rofsky, N. M. (2008). MR classification of renal masses with pathologic correlation. European Radiology, 18(2), 365–375. https://doi.org/10.1007/s00330-007-0757-0
Rini, B. I., Campbell, S. C., & Escudier, B. (2009). Renal cell carcinoma. The Lancet, 373(9669), 1119–1132. https://doi.org/10.1016/S0140-6736(09)60229-4
Rosenkrantz, A. B., Niver, B. E., Fitzgerald, E. F., Babb, J. S., Chandarana, H., & Melamed, J. (2010). Utility of the Apparent Diffusion Coefficient for Distinguishing Clear Cell Renal Cell Carcinoma of Low and High Nuclear Grade. American Journal of Roentgenology, 195(5), W344–W351. https://doi.org/10.2214/AJR.10.4688
Rossi, S. H., Prezzi, D., Kelly-Morland, C., & Goh, V. (2018). Imaging for the diagnosis and response assessment of renal tumours. World Journal of Urology, 36(12), 1927–1942. https://doi.org/10.1007/s00345-018-2342-3
Roy, C., El Ghali, S., Buy, X., Lindner, V., Lang, H., Saussine, C., & Jacqmin, D. (2005). Significance of the Pseudocapsule on MRI of Renal Neoplasms and Its Potential Application for Local Staging: A Retrospective Study. American Journal of Roentgenology, 184(1), 113–120. https://doi.org/10.2214/ajr.184.1.01840113
Salman, R. A.-S., Beller, E., Kagan, J., Hemminki, E., Phillips, R. S., Savulescu, J., Macleod, M., Wisely, J., & Chalmers, I. (2014). Increasing value and reducing waste in biomedical research regulation and management. The Lancet, 383(9912), 176–185. https://doi.org/10.1016/S0140-6736(13)62297-7
Sudharson, S., & Kokil, P. (2020). An ensemble of deep neural networks for kidney ultrasound image classification. Computer Methods and Programs in Biomedicine, 197, 105709. https://doi.org/10.1016/j.cmpb.2020.105709
Sun, M. R. M., Ngo, L., Genega, E. M., Atkins, M. B., Finn, M. E., Rofsky, N. M., & Pedrosa, I. (2009). Renal Cell Carcinoma: Dynamic Contrast-enhanced MR Imaging for Differentiation of Tumor Subtypes—Correlation with Pathologic Findings. Radiology, 250(3), 793–802. https://doi.org/10.1148/radiol.2503080995
Sun, M., Thuret, R., Abdollah, F., Lughezzani, G., Schmitges, J., Tian, Z., Shariat, S. F., Montorsi, F., Patard, J.-J., Perrotte, P., & Karakiewicz, P. I. (2011). Age-Adjusted Incidence, Mortality, and Survival Rates of Stage-Specific Renal Cell Carcinoma in North America: A Trend Analysis. European Urology, 59(1), 135–141. https://doi.org/10.1016/j.eururo.2010.10.029
Sun, X.-Y., Feng, Q.-X., Xu, X., Zhang, J., Zhu, F.-P., Yang, Y.-H., & Zhang, Y.-D. (2020). Radiologic-Radiomic Machine Learning Models for Differentiation of Benign and Malignant Solid Renal Masses: Comparison With Expert-Level Radiologists. American Journal of Roentgenology, 214(1), W44–W54. https://doi.org/10.2214/AJR.19.21617
Tanaka, T., Huang, Y., Marukawa, Y., Tsuboi, Y., Masaoka, Y., Kojima, K., Iguchi, T., Hiraki, T., Gobara, H., Yanai, H., Nasu, Y., & Kanazawa, S. (2020). Differentiation of Small (<= 4 cm) Renal Masses on Multiphase Contrast-Enhanced CT by Deep Learning. Am J Roentgenol, 214(3), 605–612. https://doi.org/10.2214/AJR.19.22074
Toda, N., Hashimoto, M., Arita, Y., Haque, H., Akita, H., Akashi, T., Gobara, H., Nishie, A., Yakami, M., Nakamoto, A., Watadani, T., Oya, M., & Jinzaki, M. (2022). Deep Learning Algorithm for Fully Automated Detection of Small (≤4 cm) Renal Cell Carcinoma in Contrast-Enhanced Computed Tomography Using a Multicenter Database. Investigative Radiology, 57(5), 327–333. https://doi.org/10.1097/RLI.0000000000000842
Weber, M. (1946). Science as Vocation. In H. H. Gerth & C. W. Mills, From Max Weber. Free press.
Xi, I. L., Zhao, Y., Wang, R., Chang, M., Purkayastha, S., Chang, K., Huang, R. Y., Silva, A. C., Vallières, M., Habibollahi, P., Fan, Y., Zou, B., Gade, T. P., Zhang, P. J., Soulen, M. C., Zhang, Z., Bai, H. X., & Stavropoulos, S. W. (2020). Deep Learning to Distinguish Benign from Malignant Renal Lesions Based on Routine MR Imaging. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 26(8), 1944–1952. https://doi.org/10.1158/1078-0432.CCR-19-0374
Xia, K., Yin, H., & Zhang, Y. (2019). Deep Semantic Segmentation of Kidney and Space-Occupying Lesion Area Based on SCNN and ResNet Models Combined with SIFT-Flow Algorithm. Journal of Medical Systems, 43(1), 2. https://doi.org/10.1007/s10916-018-1116-1
Xu, Q., Zhu, Q., Liu, H., Chang, L., Duan, S., Dou, W., Li, S., & Ye, J. (2022). Differentiating Benign from Malignant Renal Tumors Using T2-and Diffusion-Weighted Images: A Comparison of Deep Learning and Radiomics Models Versus Assessment from Radiologists. Journal of Magnetic Resonance Imaging, 55(4), 1251–1259. https://doi.org/10.1002/jmri.27900
Yang, G., Wang, C., Yang, J., Chen, Y., Tang, L., Shao, P., Dillenseger, J.-L., Shu, H., & Luo, L. (2020). Weakly-supervised convolutional neural networks of renal tumor segmentation in abdominal CTA images. Bmc Medical Imaging, 20(1), 37. https://doi.org/10.1186/s12880-020-00435-w
Yang, Y., Xie, L., Zheng, J.-L., Tan, Y.-T., Zhang, W., & Xiang, Y.-B. (2013). Incidence Trends of Urinary Bladder and Kidney Cancers in Urban Shanghai, 1973-2005. PLoS ONE, 8(12), e82430. https://doi.org/10.1371/journal.pone.0082430
Young, J. R., Margolis, D., Sauk, S., Pantuck, A. J., Sayre, J., & Raman, S. S. (2013). Clear Cell Renal Cell Carcinoma: Discrimination from Other Renal Cell Carcinoma Subtypes and Oncocytoma at Multiphasic Multidetector CT. Radiology, 267(2), 444–453. https://doi.org/10.1148/radiol.13112617
Zabihollahy, F., Schieda, N., Krishna, S., & Ukwatta, E. (2020). Automated classification of solid renal masses on contrast-enhanced computed tomography images using convolutional neural network with decision fusion. European Radiology, 30(9), 5183–5190. https://doi.org/10.1007/s00330-020-06787-9
Zhang, J., Lefkowitz, R. A., Ishill, N. M., Wang, L., Moskowitz, C. S., Russo, P., Eisenberg, H., & Hricak, H. (2007). Solid Renal Cortical Tumors: Differentiation with CT. Radiology, 244(2), 494–504. https://doi.org/10.1148/radiol.2442060927
Zhao, Y., Chang, M., Wang, R., Xi, I. L., Chang, K., Huang, R. Y., Vallières, M., Habibollahi, P., Dagli, M. S., Palmer, M., Zhang, P. J., Silva, A. C., Yang, L., Soulen, M. C., Zhang, Z., Bai, H. X., & Stavropoulos, S. W. (2020). Deep Learning Based on MRI for Differentiation of Low- and High-Grade in Low-Stage Renal Cell Carcinoma. Journal of Magnetic Resonance Imaging: JMRI, 52(5), 1542–1549. https://doi.org/10.1002/jmri.27153
Zheng, Y., Wang, S., Chen, Y., & Du, H.-Q. (2021). Deep learning with a convolutional neural network model to differentiate renal parenchymal tumors: A preliminary study. Abdominal Radiology (New York), 46(7), 3260–3268. https://doi.org/10.1007/s00261-021-02981-5
Zhou, Y., & Volkwein, J. F. (2004). Examining the Influences on Faculty Departure Intentions: A Comparison of Tenured Versus Nontenured Faculty at Research Universities Using NSOPF-99. Research in Higher Education, 45(2), 139–176. https://doi.org/10.1023/B:RIHE.0000015693.38603.4c
Zhu, X.-L., Shen, H.-B., Sun, H., Duan, L.-X., & Xu, Y.-Y. (2022). Improving segmentation and classification of renal tumors in small sample 3D CT images using transfer learning with convolutional neural networks. International Journal of Computer Assisted Radiology and Surgery, 17(7), 1303–1311. https://doi.org/10.1007/s11548-022-02587-2
Znaor, A., Lortet-Tieulent, J., Laversanne, M., Jemal, A., & Bray, F. (2015). International Variations and Trends in Renal Cell Carcinoma Incidence and Mortality. European Urology, 67(3), 519–530. https://doi.org/10.1016/j.eururo.2014.10.002